Technological Breakthrough: Membrane Proteomics to Analyze Cell Membrane Signal Transduction
Online InquiryDue to the development of sophisticated tools in membrane proteomics, the study of cell membrane signal transduction has made notable advancements in the dynamic field of biological research. These innovative techniques, such as phosphorylated protein analysis and studies of membrane protein complexes, have revealed previously unattainable insights into the complicated mechanisms driving cellular communication.
Phosphorylated Protein Analysis: Unraveling the Intricacies of Cell Membrane Signaling
Cell membrane signaling is a sophisticated process driven by intricate protein interactions and post-translational modifications. Among these modifications, phosphorylation stands out as a pivotal regulatory mechanism, governing the activation or inhibition of key signaling pathways.
The Significance of Phosphorylation
Phosphorylation, the addition of a phosphate group to a protein molecule, is a reversible modification that acts as a molecular switch, dictating protein activity and function. Cell membrane receptors, often spanning the lipid bilayer, play a vital role in sensing extracellular cues and initiating intracellular responses. The phosphorylation of specific residues within these receptors triggers downstream signaling cascades, transmitting information across the cell membrane and eliciting cellular responses.
Advanced Techniques: Mass Spectrometry and Liquid Chromatography
Creative Proteomics harnesses state-of-the-art techniques like Mass Spectrometry (MS) and Liquid Chromatography (LC) to dissect phosphorylated protein networks. MS allows for the identification and quantification of phosphorylated peptides, enabling the comprehensive profiling of phosphorylation events. Complex peptide mixtures can be better separated using LC, which helps to precisely identify phosphorylation sites and their occupancy.
Mapping the Phosphoproteome
Through phosphoproteomic analysis, researchers can map the entire phosphoproteome of a cell or tissue. This high-throughput approach enables the identification of hundreds to thousands of phosphorylation sites across various proteins. These sites often serve as molecular footprints of signal transduction pathways, pointing towards key players and intricate regulatory nodes.
Case Study: Phosphorylated Protein Analysis
Background
Phosphoproteomics is a critical area of research focused on understanding cellular signaling networks by identifying and quantifying phosphorylation sites and mapping the enzymes involved in these post-translational modifications (PTMs). Cellular signaling networks play a crucial role in various biological processes, and the dysregulation of phosphorylation events is associated with diseases like cancer. Researchers use multiple analytical methods to identify proteins and protein phosphorylation sites that regulate cell signaling and to quantify the dynamic responses of these sites to different cellular stimuli(White FM, et al. 2016).
Sample
The samples for phosphoproteomics studies typically involve cellular lysates or tissues. These samples are treated or stimulated under specific conditions to activate signaling pathways. The complexity of the sample, including the number of proteins and PTMs, poses a significant challenge for analysis.
Technical Approach
Several methods are employed to investigate phosphorylation events and enzyme-substrate relationships in cellular signaling networks:
- Enzyme–Substrate Mapping: In vitro enzyme assays and protein microarrays are used to test the ability of enzymes (kinases/phosphatases) to interact with known or potential substrates. These assays provide direct evidence of enzyme-substrate interactions but are often low throughput and may not fully mimic cellular conditions.
- Chemical Genetics: This approach involves modifying kinases to create analog-sensitive (AS) kinases, allowing the use of bio-orthogonal ATP analogs to phosphorylate substrates. SILAC is used to distinguish substrate phosphorylation by AS kinases from background labeling. Membrane permeabilization techniques help overcome ATP analog impermeability.
- Kinase Activation and Inhibition: Cells are stimulated in the presence or absence of kinase agonists or inhibitors. Global untargeted phosphoproteomics is employed to identify potential substrates. However, this approach can lead to complex responses in signaling networks, making it challenging to distinguish direct substrates from indirect ones.
- Substrate-Enzyme Mapping: Motif matching is used to predict the kinases responsible for phosphorylation sites based on amino acid sequences. Chemical genetics for serine/threonine phosphorylation sites involves converting phosphorylated residues to cysteines and employing chemical approaches to identify the kinases involved.
Results
Phosphoproteomics studies aim to uncover phosphorylation sites, quantify their changes in response to stimuli, and map the enzymes responsible for these PTMs. Results often include lists of identified phosphorylation sites, changes in phosphorylation levels under different conditions, and potential substrates for specific kinases. These findings provide critical insights into cellular signaling networks and can help elucidate mechanisms underlying cellular responses to stimuli and diseases such as cancer. Data quality is a paramount concern, and results are typically validated through manual spectral analysis, peptide standards, co-elution studies, and orthogonal assays to ensure accuracy and reliability.
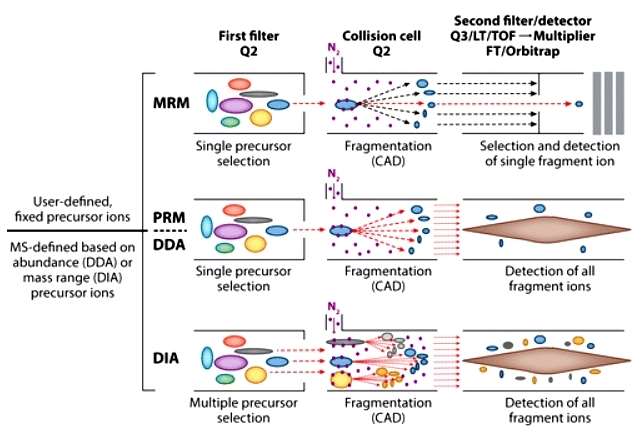 A schematic of how peptides are isolated, fragmented, and analyzed (White FM, et al,. 2016)
A schematic of how peptides are isolated, fragmented, and analyzed (White FM, et al,. 2016)
Case Study: Studying Membrane Protein Complexes
Background
The study focuses on understanding the role of transthyretin (TTR) in regulating the function of STRA6, a plasma membrane protein involved in vitamin A metabolism and signaling. Specifically, the researchers investigate whether TTR influences the uptake of retinol (vitamin A) by STRA6 and its subsequent signaling activities. Vitamin A plays critical roles in various biological processes, and its transport in the bloodstream is facilitated by retinol-binding protein (RBP), which is often complexed with TTR (Berry DC, et al. 2012).
Sample
The study employs various experimental models, including cell lines such as HepG2, NIH 3T3, and NIH 3T3-L1, as well as animal models (mice) with a mixed genetic background. Recombinant proteins, mutant RBP, and chemical cross-linkers are used in the experiments. The researchers also make use of glucose tolerance tests (GTT) to assess insulin responses in mice.
Technical Methods
- Cell Culture: HepG2, NIH 3T3, and NIH 3T3-L1 cells are cultured in specific media with serum supplements.
- RNA Analysis: RNA is extracted from cells, and cDNA is generated for quantitative PCR (Q-PCR) analysis of gene expression, including genes related to STRA6 signaling.
- Recombinant Proteins: Recombinant RBP and TTR are produced using bacterial expression vectors and purified for use in experiments.
- Vitamin A Uptake Assays: Cellular uptake of retinol is assessed by incubating cells with labeled holo-RBP or holo-RBP-TTR complexes. The rate of uptake is measured using scintillation counting, and data are normalized based on protein content.
- In Vivo Studies: Mice are used to investigate the effects of RBP, TTR, and their complexes on vitamin A uptake and insulin responses. Intraperitoneal injections of labeled proteins are administered, and tissues are analyzed for radio-labeled retinol uptake.
- Immunoblots and Immunoprecipitations:Protein samples are subjected to immunoblotting and immunoprecipitation techniques to detect phosphorylation events and protein interactions.
- Fluorescence and Fluorescence Anisotropy Titrations: These methods are used to examine the binding of retinol to RBP or its mutant.
- Insulin Signaling Assays: Insulin-induced phosphorylation of the insulin receptor and downstream effector AKT is assessed in the presence of RBP, TTR, or their complexes.
Results
- TTR inhibits STRA6-mediated uptake of retinol from holo-RBP, and this inhibition is dose-dependent.
- TTR specifically blocks STRA6-mediated transport of retinol, suggesting that STRA6 associates only with free holo-RBP and not the holo-RBP-TTR complex.
- TTR also inhibits STRA6-mediated cell signaling, including phosphorylation of STAT proteins and expression of STAT target genes. This inhibition affects insulin responses in cell cultures.
- In vivo studies in mice demonstrate that TTR prevents holo-RBP-induced insulin resistance when administered in conjunction with holo-RBP, highlighting the physiological relevance of these findings.
Additionally, the researchers observe that the RBP/TTR ratio in the bloodstream, which can be altered in obesity, is crucial for regulating STRA6 signaling. Elevated RBP levels in obese individuals may lead to increased RBP/TTR ratios, potentially impacting STRA6-mediated processes.
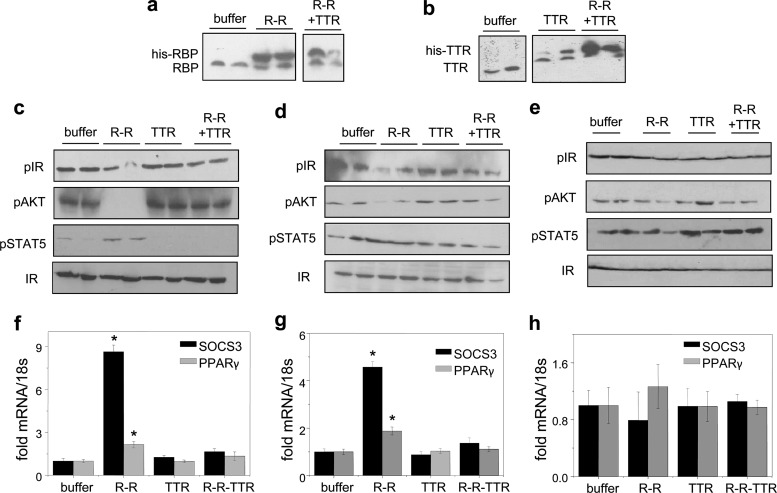 TTR suppresses activation of STRA6 by holo-RBP in vivo (Berry DC, et al,. 2012)
TTR suppresses activation of STRA6 by holo-RBP in vivo (Berry DC, et al,. 2012)
Learn more
In conclusion, the breakthroughs achieved through membrane proteomics have revolutionized the study of cell membrane signal transduction. Creative Proteomics' role in advancing these techniques underscores its commitment to pushing the boundaries of biological research. As technology continues to evolve, the partnership between Creative Proteomics and cutting-edge membrane proteomics promises a future where the complexities of cellular communication are fully understood, unlocking new avenues for therapeutic interventions.
Creative Proteomics offer comprehensive membrane proteomics research services including membrane proteomics analysis, membrane lipidomic analysis, membrane Protein-molecule interaction analysis, membrane protein structure determination, among others. We play an important role in advancing membrane proteomics research.
References
- White FM, Wolf-Yadlin A. Methods for the Analysis of Protein Phosphorylation-Mediated Cellular Signaling Networks. Annu Rev Anal Chem (Palo Alto Calif). 2016;9 (1):295-315.
- Berry DC, Croniger CM, Ghyselinck NB, Noy N. Transthyretin blocks retinol uptake and cell signaling by the holo-retinol-binding protein receptor STRA6. Mol Cell Biol. 2012 ;32(19):3851-9.
Related Services
* For Research Use Only. Not for use in diagnostic procedures.