Nucleolar Proteomics Services
Online InquiryThe nucleolus is the most prominent structural and functional nuclear compartment of eukaryotic cells. The main function of the nucleoli is to serve as a site for rDNA transcription, processing, and assembly of large and small ribosomes in eukaryotic cells. The nucleolus also has other functions not directly related to ribosome biosynthesis. For example, the assembly of signal recognition particles (SRP), the production and transportation of mRNA, small RNA molecule modification, RNA editing, telomere maturation, cell cycle regulation and sensing of cell stress.
In order to better analyze the progress of nucleolar structure, function, proteomics and its origin and evolution, Creative Proteomics provides a one-stop service for nucleolar proteomics, and conducts a comprehensive analysis of nucleolar proteomics from a holistic perspective. The main contents include: 1) Analysis and purification of nucleoli; 2) Structural analysis of nucleoli; 3) Functional analysis of nucleoli; 4) Identification and quantification of nucleoli protein; 5) Functional analysis of nucleoli protein domain; 6) Data analysis and bioinformatics analysis.
We Provide the Following Services, including but not Limited to:
Separation and purification of nucleoli
- Sucrose density gradient
- The rapid, efficient, and practical (REAP) method
Structural analysis of nucleoli
The study of three characteristic regions of the nucleolus, including the fibrillar centres (FC) in the lightly dyed low electron density region embedded in the particle component, the dense fibrillar component (DFC) with the highest electron density in the ultrastructure, and metabolically active granular component (GC). We provide differential studies of animal and plant nucleoli. In addition, we also provide correlation analysis of nucleolar structure and the rate of nuclear ribosomal biosynthesis.
Functional analysis of nucleoli
- Involved in metabolic analysis of RNA other than rRNA, such as mRNA generation and transportation, RNA editing, tRNA maturation, SRP assembly, processing and maturation of small nuclear and nucleolar small RNA, and heterochromatin siRNA pathway.
- Involved in the research of regulating cell function, including protein translation, cell cycle regulation, cell stress response process, and cell senescence.
Qualitative and quantitative analysis of nucleoli protein
Creative Proteomics analyzes nucleolar proteins between different species. Proteins are separated using common protein separation techniques, and these proteins are subjected to mass spectrometry to identify nucleolar proteins. Differential analysis of the identified proteins is provided. For quantitative analysis, both labeled and labeled-free quantification strategies are available. Label-based quantification technologies include: SILAC, TMT, and iTRAQ. Labeled-free quantitative technologies include SRM / MRM, PRM and SWATH technologies. Different quantitative techniques have their unique advantages and characteristics.
Bioinformatics analysis
For example, nucleolar protein domain function analysis, homology analysis and evolution analysis.
Advantages of the Nucleolar Proteomics Services:
- One-stop service: we will customize a solution for you based on your samples and needs. You can also choose which technique to use for analysis.
- High performance mass spectrometry system: equipped with Q Exactive Hybrid Quadrupole-Orbitrap and Agilent 6540 Q-TOF high performance mass spectrometer with multiple mass spectrometry techniques such as SRM, PRM and SWATH full scan collection, which effectively improving the identification and quantification of low abundance subcellular related proteins.
- Multi-database integration: by integrating multiple related high-quality protein databases, the accuracy and flux of subcellular protein localization can be greatly improved.
- Multi-dimensional data analysis: in addition to the comparison with the more complete related subcellular database, multiple software was used for the prediction analysis of subcellular localization based on the calculation method to further increase the identification flux.
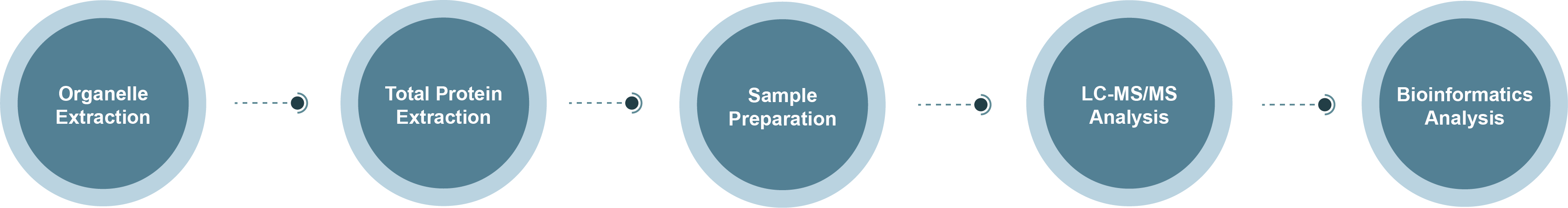
Nucleolar Proteomics Technology Roadmap
Data Analysis of Nucleolar Proteomics
| 1. Data output statistics and quality control | 2. Protein identification analysis (Density map, Scatter plot, Venn diagram) | 3. Quantitative protein analysis (Density map, Scatter plot, Venn diagram) |
| 4. Protein GO analysis | 5. Protein pathway analysis | 6. Protein COG analysis |
| 7. GO enrichment analysis of differential proteins | 8. Pathway enrichment analysis of differential proteins | 9. COG analysis of differential proteins |
References
- Montacié C, Durut N, et al. Nucleolar proteome analysis and proteasomal activity assays reveal a link between nucleolus and 26S proteasome in A. thaliana. Frontiers in plant science, 2017, 8: 1815.
- Nicolas A, Bensaddek D. Analysis of Mass Spectrometry Data for Nucleolar Proteomics Experiments//The Nucleolus. Humana Press, New York, NY, 2016: 263-276.
- Andersen J S, Lam Y W, et al. Nucleolar proteome dynamics. Nature, 2005, 433(7021): 77-83.
Related Services
* For Research Use Only. Not for use in diagnostic procedures.