Peroxisome Proteomics Services
Online InquiryPeroxisomes are widely found in almost all animal and plant cells except red blood cells. It is wrapped in a monolayer of metabolic enzymes and contains no DNA components. Its main function is to participate in lipid metabolism. In addition, peroxisomes inactivate toxic substances, such as hydrogen peroxide, to protect cells from these toxins. If the number of peroxisomes in the cell is reduced or completely absent, its function will be abnormal and lead to disease.
Creative Proteomics provides a one-stop service for peroxisome proteomics, which aims to conduct a comprehensive analysis of peroxisome from a holistic perspective. The main contents include: 1) Study on the biogenesis of peroxisomes; 2) Study the functions of membrane proteins and matrix proteins of peroxisomes;3) Studying the links between peroxisomes and other diseases.
We Provide the Following Services, including but not Limited to:
- Peroxisome in animals
- Study the biogenesis of peroxidase
- Analysis of its role in fatty acid oxidation
- Study the relationship between peroxidase and Alzheimer's disease, neurodegenerative disease, and peroxisomal defects, et al.
- Peroxisomes in plants
- Study the biogenesis of peroxidase
- Study the relationship between peroxidase protein and photorespiration
- Study the relationship between oxidation of peroxidase protein and fat and glyoxylic acid cycle
Metabolic enzymes: mainly involved in the removal of peroxides and the decomposition and synthesis of lipids, etc.
Constitutive protein: it is called peroxin (PEX), and is involved in the peroxisome biogenesis, division and proliferation.
Advantages of the Peroxisome Proteomics Service:
- Optimized protein extraction strategies: the extraction scheme of conventional peroxidase protein is optimized to increase the extraction purity by 10-20%.
- High performance mass spectrometry system: equipped with Q Exactive Hybrid Quadrupole-Orbitrap and Agilent 6540 Q-TOF high performance mass spectrometer with multiple mass spectrometry techniques such as SRM, PRM and SWATH full scan collection, which effectively improving the identification and quantification of low abundance subcellular related proteins.
- Multi-database integration: by integrating multiple related high-quality protein databases, the accuracy and flux of subcellular protein localization can be greatly improved.
- Multi-dimensional data analysis: in addition to the comparison with the more complete related subcellular database, multiple software was used for the prediction analysis of subcellular localization based on the calculation method to further increase the identification flux.
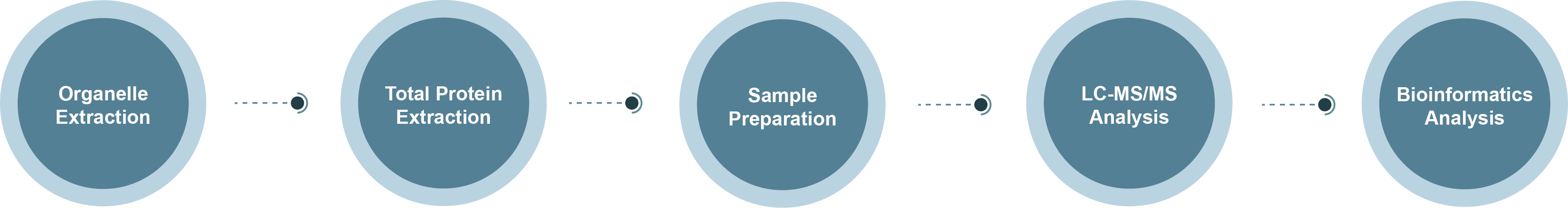
Peroxisome Proteomics Technology Roadmap
Data Analysis of Peroxisome Proteomics
| 1. Data output statistics and quality control | 2. Protein identification analysis (Density map, Scatter plot, Venn diagram) | 3. Quantitative protein analysis (Density map, Scatter plot, Venn diagram) |
| 4. Protein GO analysis | 5. Protein pathway analysis | 6. Protein COG analysis |
| 7. GO enrichment analysis of differential proteins | 8. Pathway enrichment analysis of differential proteins | 9. COG analysis of differential proteins |
References
- Montserrat A, Kersten S. Regulation of lipid droplet-associated proteins by peroxisome proliferator-activated receptors. Biochimica et Biophysica Acta (BBA)-Molecular and Cell Biology of Lipids, 2017, 1862(10): 1212-1220.
- Tripathi D N, Walker C L. The peroxisome as a cell signaling organelle. Current opinion in cell biology, 2016, 39: 109-112.
- Baker A, Hogg T L, Warriner S L. Peroxisome protein import: a complex journey. Biochemical Society Transactions, 2016, 44(3): 783-789.
Related Services
* For Research Use Only. Not for use in diagnostic procedures.