SILAC Quantitation Service
Online InquiryStable isotope labeling with amino acids in cell culture (SILAC) is a widely used in vivo labeling technique. This method can identify and quantify the relative changes of the target protein, and provides an effective method for the comprehensive and systematic qualitative and quantitative analysis of proteomes of complex mammalian cells.
Creative Proteomics has extensive experience with label-based quantitative proteomics. We can provide in vivo labeled SILAC quantitation service to help you perform quantitative analysis of proteomics.
SILAC Quantitation Technology Principle and Process
The basic principle of SILAC is to replace corresponding amino acids in cell culture medium with essential amino acids labeled with natural isotopes (light) or stable isotopes (heavy). After 5-6 doubling cycles of the cell, the stable isotope-labeled amino acid is completely incorporated into the newly synthesized protein of the cell, replacing the original amino acid. Lytic proteins of different labeled cells are mixed in proportion to the number of cells or the amount of proteins, and then identified by mass spectrometry after separation and purification.
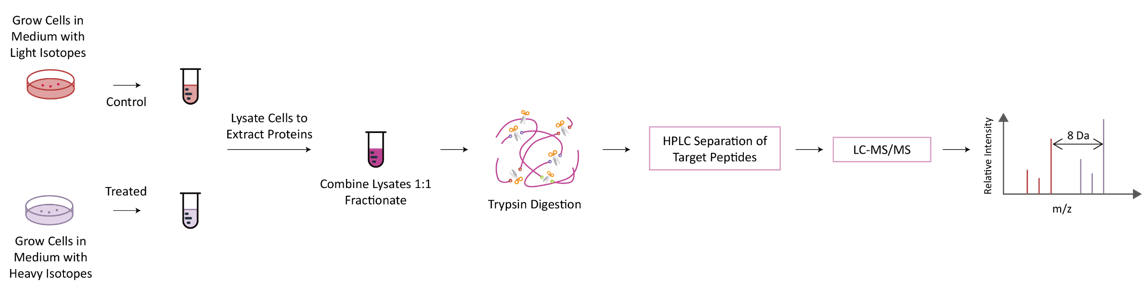
Technology Platform
Q Exactive Hybrid Quadrupole-Orbitrap and Agilent 6540 Q-TOF high performance mass spectrometer.
Characteristics of SILAC Quantitation Service:
- In vivo labeling technology, closer to the true state of the sample. Stable isotope-labeled amino acids are basically the same as natural amino acids in chemical properties, and there is almost no difference in biological behavior between labeled cells and unlabeled cells. Labeling efficiency can be as high as 100%.
- Combined with mass spectrometry for quantitative analysis, the quantitative results are accurate and repeatable.
- In vivo metabolic labeling combined with SDS-PAGE or chromatographic separation technology can be compatible with hydrophobic and basic proteins without being restricted by protein properties.
- Simultaneous separation, digestion, and identification of multiple samples after mixing, reducing the impact of subsequent operations and equipment.
- SILAC requires a small amount of protein compared to chemical markers, typically just a few dozen micrograms per sample.
- It is only suitable for cells cultured in vivo, and cannot be analyzed for tissue samples and body fluid samples commonly used in biomedical research. The labeling cost of animal models is too high to implement.
- Multiple enrichment techniques can be combined to help detect changes in the abundance of low-abundance proteins and post-translational modifications.
Application of SILAC Quantitative Technology
- Comparison of proteins in multiple samples.
- Comparison of proteins in the same sample under different conditions.
- Combining SILAC with IP technology can be used for protein interaction analysis.
- Through the interpretation of the mass spectrum, confirmation of phosphorylation sites and quantitative analysis of phosphorylated proteins can be performed.
Delivery
Experimental protocols, raw data, mass spectrogram, and bioinformatics analysis report. Reports are available in Excel or PDF format.
Turnaround Time
Typical 40 working days from sample QC acceptance to data report delivery.
Want to Know about Other Labeled Proteomics Quantitative Analysis Techniques?
- SILAC Quantitation Service
- ICAT Quantitation Service
- TMT Quantitation Service
- iTRAQ Quantification Service
- SRM/MRM Quantitation Service
- PRM Quantitation Service
References
- Chen X, Wei S, Ji Y, et al. Quantitative proteomics using SILAC: Principles, applications, and developments. Proteomics, 2015, 15(18): 3175-3192.
- Güther M L S, Urbaniak M D, Tavendale A, et al. High-confidence glycosome proteome for procyclic form Trypanosoma brucei by epitope-tag organelle enrichment and SILAC proteomics. Journal of proteome research, 2014, 13(6): 2796-2806.
* For Research Use Only. Not for use in diagnostic procedures.