iTRAQ Quantification Service
Online Inquiry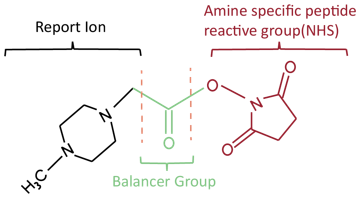
Isobaric tags for relative and absolute quantitation (iTRAQ) is an effective way to study the different physiological states of cells or tissues, or compare the similarities and differences between proteomes in normal and pathological states. It can realize the quantification of proteins in up to 8 different samples at one time, which is a high-throughput screening technology commonly used in quantitative proteomics.
Creative Proteomics provides iTRAQ quantitative services, which can accurately quantify and identify whole proteins or proteins in complex mixed systems in organelles, accelerating identification and screening of differentially expressed proteins between samples and qualitative and quantitative analysis of key proteins. Combined with bioinformatics analysis, the physiological and pathological functions of cells can be revealed.
iTRAQ Quantitation Technology Principle and Process
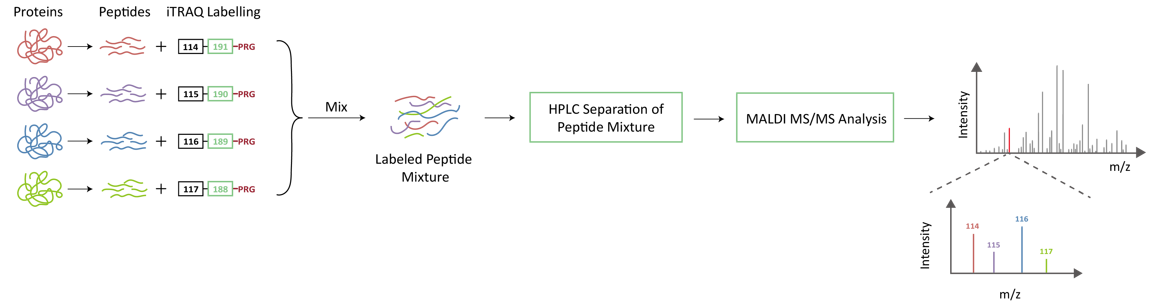
iTRAQ usually consists of a reporter group, a balancing group and a peptide reaction group. The mass sum of different reporting groups and their corresponding balancing groups are the same. The reactive group can be attached to the amino terminus of all peptide chains for labeling. In primary mass spectrometry, peptides with different labels have the same mass charge ratio and therefore appear in the same mass spectrum peak. Upon entry to secondary mass spectrometry, the reporter group dissociates to produce an ionic signal that provides quantitative information about the peptides in the sample.
Technology Platform
Q Exactive Hybrid Quadrupole-Orbitrap and Agilent 6540 Q-TOF high performance mass spectrometers are available.
Characteristics of iTRAQ Quantitation Service:
- High Throughput: Up to 8 sample proteins can be identified and quantified simultaneously in a single run. More than 8 samples can be bridged by setting internal parameters to achieve multiple comparisons on the machine data.
- Accurate quantification and high repeatability: Provide a high-resolution mass spectrometer platform. Different samples are mixed after being labeled and processed in a unified manner, which reduces the experimental errors caused by sample processing and computer operation.
- Wide range of protein detection: all known proteins expressed in the sample can be detected, such as high molecular weight proteins, acid-base proteins, cytosolic proteins, membrane proteins, nuclear proteins, extracellular proteins, secreted proteins and insoluble proteins. Large span of protein abundance and molecular mass, low abundance proteins, strong basic proteins, proteins less than 10KD or more than 200KD can be detected.
- Support correlation analysis: Comprehensive experimental data such as upstream transcriptomes are provided to provide multi-omics correlation analysis.
Applications of iTRAQ Quantitative Technology
- Screening for disease markers
- Specific functional protein screening
- Drug target research
Sample Requirements
- Protein samples: concentration >1μg/μL, total protein >100μg
- Cell samples: cell volume>107
- Animal tissue: >1g
- Plant tissue: >200mg
Delivery
Experimental procedures, liquid chromatography and mass spectrometer parameters, MS raw data files, bioinformatics analysis charts. Reports are available in Excel or PDF format.
Bioinformatics analysis includes:
- Statistical analysis of proteins: Venn diagram, volcano map
- Functional annotations: GO annotations, KEGG annotations, COG annotations
- Cluster analysis: hierarchical clustering, K-means clustering
- Network analysis: STRING analysis
Want to Know about Other Labeled Proteomics Quantitative Analysis Techniques?
- SILAC Quantitation Service
- ICAT Quantitation Service
- TMT Quantitation Service
- iTRAQ Quantification Service
- SRM/MRM Quantitation Service
- PRM Quantitation Service
References
- Christoforou A, Arias A M, Lilley K S. Determining protein subcellular localization in mammalian cell culture with biochemical fractionation and iTRAQ 8-plex quantification//Shotgun Proteomics. Humana Press, New York, NY, 2014: 157-174.
- Lilley K S, Dupree P. Plant organelle proteomics. Current opinion in plant biology, 2007, 10(6): 594-599.
- Sun Y, Hu B, Fan C, et al. iTRAQ-based quantitative subcellular proteomic analysis of Avibirnavirus-infected cells. Electrophoresis, 2015, 36(14): 1596-1611.
* For Research Use Only. Not for use in diagnostic procedures.