Isotope-Coded Affinity Tag Protein Quantitation Service
Online InquiryCreative Proteomics has an isotope-coded affinity tags (ICATs) and tandem mass spectrometry platform to quantify changes in protein expression in proteomics studies.
Isotope-coded affinity tag (ICAT) technology, as a relative quantitative method for labeling stable isotopes in vitro, has become an important proteomics quantitative analysis scheme. A stable isotope-encoded affinity tag can be used as an affinity reagent for chemical synthesis capable of reacting with cysteine. Isotope-coded affinity tag technology can be used in vitro to label with both light and heavy (stable heavy isotope) forms of affinity tags. The labeled peptides can be digested with trypsin, and the labeled peptides are separated and purified, and then analyzed by mass spectrometry. Similar to the in vivo labeling method, paired peaks can be obtained which can indicate the peptides and corresponding protein content in different samples. This stable isotope affinity tagging technology can be widely used in quantitative proteomics analysis of cells and tissues, providing accurate protein relative quantitative data.
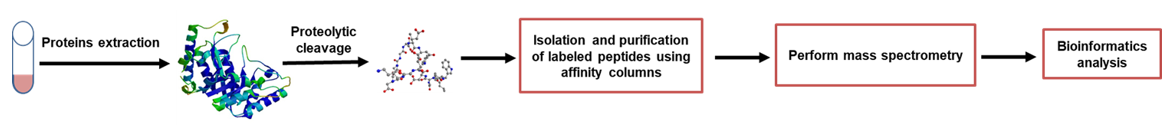
The Workflow of Isotope-Coded Affinity Tag Protein Quantitation Service
The process for isotope-coded affinity tag protein quantitation technology includes protein extraction, proteolytic cleavage, isolation and purification of labeled peptides using affinity columns, perform mass spectrometry, analyze the relative protein quantitative data, and bioinformatics analysis.
Technology Platform
Agilent 6125B Single Quadrupole LC/MS.
Advantages of Isotope-Coded Affinity Tag Protein Quantitation Service:
- The isotope-coded affinity labeling (ICAT) technology eliminates the need for complicated biphasic gel electrophoresis, which increases the direct capture and reliability of low-abundance proteins, especially hydrophobic proteins such as membrane proteins.
- ICAT technology simplifies the detection process, allowing direct testing of mixed samples from normal and diseased cells or tissue.
- It can quickly find important functional proteins such as disease-related proteins and biomarkers, which can be quickly used for disease diagnosis.
- It uses an acid-sensitive linker, which reduces the relative molecular mass added to the peptide and improves the coverage of proteins detected by mass spectrometry.
- This technology is not radioactive, no special protection is required in the process of separation, labeling compound synthesis and application, which is safety and non-toxic and can be directly used in many fields such as animals.
- It is compatible with the analysis of most proteins in body fluids, cells and tissues under any conditions.
- ICAT technology allows any type of biochemical, immunological, and physical separation methods, so it can be used to quantify trace proteins well.
Application of Isotope-Coded Affinity Tag Protein Quantitative Technology
- Quantitative proteomics analysis of cells and tissues.
- Clinical research and diagnosis of various diseases in the medical field.
- Detecting ecological processes that change over time and space in environmental science research.
- Human nutrition in energy metabolism research.
- Nutrition metabolism in glycolipid metabolism.
- Research on the improvement of agricultural products and other crops in agricultural scientific research.
- For the analysis of food, pesticide residues, stimulants, heroin, etc. in the field of analytical testing.
Delivery
Target protein candidate peptide quantitative results, mass spectrogram, and bioinformatics analysis report.
Want to Know about Other Subcellular Proteomics Quantitative Analysis Techniques?
- SILAC Quantitation Service
- ICAT Quantitation Service
- TMT Quantitation Service
- iTRAQ Quantification Service
- SRM/MRM Quantitation Service
- PRM Quantitation Service
References
- Jiang XS, Dai J, Sheng QH, et al. A comparative proteomic strategy for subcellular proteome research: ICAT approach coupled with bioinformatics prediction to ascertain rat liver mitochondrial proteins and indication of mitochondrial localization for catalase. Mol Cell Proteomics, 2005, 4:12-34.
- Shi Y, Xiang R, Crawford JK, et al. A simple solid phase mass tagging approach for quantitative proteomics. J Proteome Res, 2004,3(1):104.
* For Research Use Only. Not for use in diagnostic procedures.