Membrane Lipidomics Service
Online InquiryLipids are an important component of cell membrane structure and play an important role in cell development, energy reserves, signal transduction, substance transport and other life activities. Therefore, the dynamic changes of lipid metabolism can directly or indirectly reflect the physiological and pathological changes of the organism.
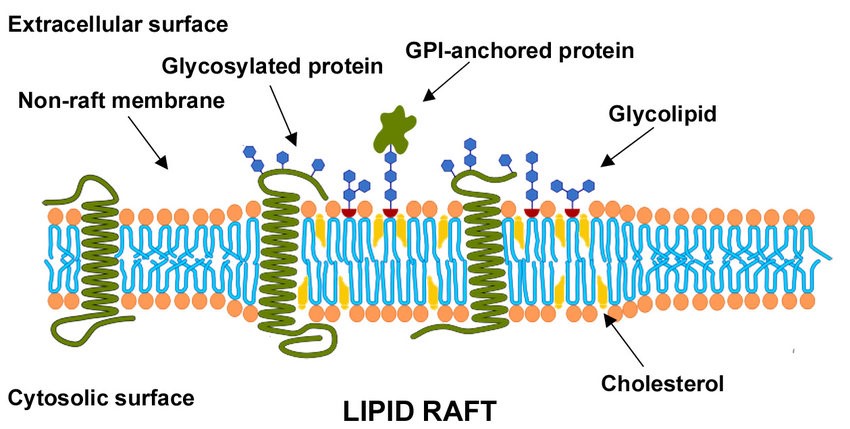 Lipid microdomains (Scolari, 2009)
Lipid microdomains (Scolari, 2009)
Creative Proteomics aims to provide high-quality membrane lipidomics services for customers in different fields. Our scientists help speed up your project progress based on professional knowledge and high-sensitivity equipment.
What we can provided:
- Targeted lipid absolute quantification
- Fatty acid composition analysis
- Metabolic regulation of lipid components in cell membranes
- Analysis of molecular composition of membrane lipids
- Differential lipid analysis under endogenous and exogenous stimuli such as changes in hormone levels, developmental changes, and environmental stresses
- Lipid biomarker screening
| Fatty acids (e.g. linoleic acid, arachidonic acid) | Glycerolipids (e.g. TG, DG, etc.) | Glycerophospholipids (e.g. PC, PE, PG, PA, etc.) |
| Sphingolipids (e.g. Cer, SM, etc.) | Solid alcohol lipids (e.g. sterol lipids) | Glycolipids (e.g. MGDG, SQDG, etc.) |
| Pregnenolone lipids (e.g. CoA, etc.) | Polyethylene esters |
Analysis strategies
- Targeted lipidomics service: Based on high-resolution mass spectrometry and isotopic internal standards, the parallel reaction monitoring (PRM) targeted analysis technique is used for the simultaneous and specific acquisition of signals from multiple lipid molecules (e.g., dozens of target lipid molecules). Targeted detection and quantification of specific lipid molecules or specific types of lipids is performed using standard products as reference. The lipid concentration is calculated from the standard curve to obtain its absolute content.
- Untargeted lipidomics services: Unbiased detection of all small molecule metabolites in as many biological samples as possible, such as cells, tissues, organs or body fluids. Capable of systematically resolving changes in lipid composition and expression in organisms, it can effectively help to study the lipid family and the alterations and functions of lipid molecules in various biological processes. Analyze the metabolic pathways of different metabolites and explore the changes of lipids in different environments.
1. Qualitative and quantitative analysis of metabolites are performed by nanoLC-MS-MS.
2. Screen different
metabolites through comparison in the control combination experiment.
3. KEGG pathway enrichment of
differential metabolites is conducted to search for key genes/enzymes for experimental verification.
4.
Correlation analysis of metabolite changes with clinical biochemical indicators and other phenotypes to support
metabolite function from another dimension.
Bioinformatics analysis
- Basic data analysis: Metabolite identification and quantitative data analysis. Composition analysis. Content difference analysis (total amount, Class level, Species level analysis)
- Advanced analysis: biomarker screening. Correlation analysis with target parameters (OPLS regression analysis, WGCNA analysis)
Creative Proteomics is based on a highly stable, reproducible and highly sensitive separation, characterization, identification and quantitative analysis system to provide you with a reliable, fast and cost-effective one-stop membrane lipidomics analysis service. Thanks for your trust. If you have any questions or need other membrane analysis related services, please contact us.
Other related services:
Reference
- Scolari, S. (2009). Lateral organization of the transmembrane domain and cytoplasmic tail of influenza virus hemagglutinin revealed by time resolved imaging.
Related Services
* For Research Use Only. Not for use in diagnostic procedures.