NTA-Based Exosome Identification
Online InquiryOver the past decades, the massive expansion of research on the biogenesis, disease relevance, and therapeutic potential of exosomes has prompted the adoption of rigorous methods to identify and characterize exosomes, involving their biophysical, phenotypic, and functional properties. To meet the growing need for accurate assessment of exosomes and to further facilitate downstream therapeutic applications, we have applied several technologies, including nanoparticle tracking analysis (NTA) for exosome identification. As an optical method, NTA can track single particles and assign sizes and counts.
NTA-based exosome identification
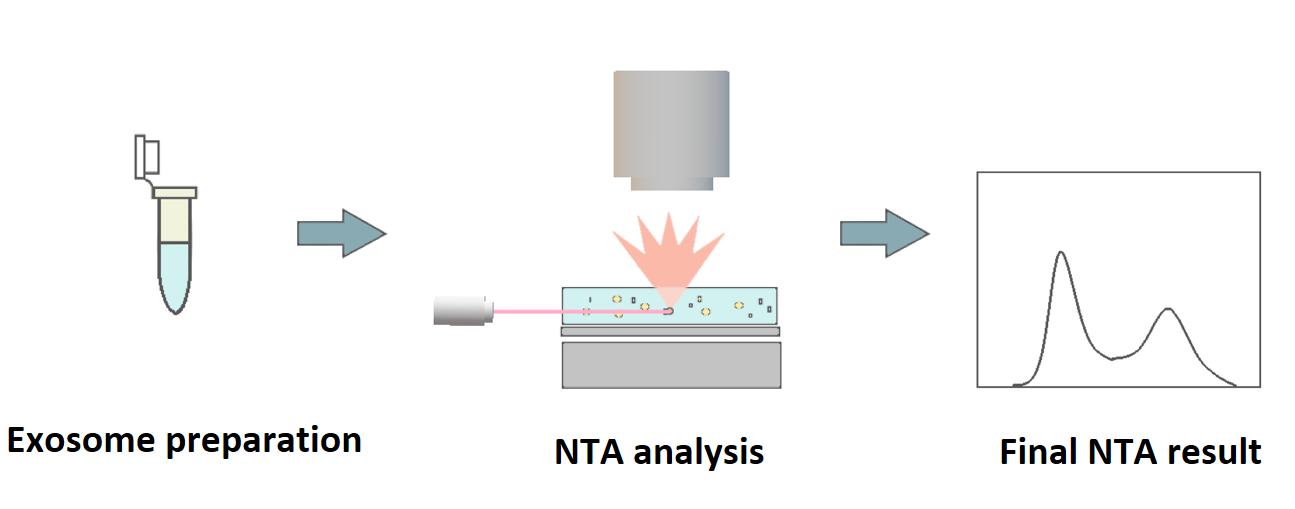
Nanoparticle tracking analysis (NTA) is a widely used and sensitive technology for the identification and analysis of particle size distribution of extracellular vesicles (EVs), including exosomes. This method is based on the characteristic movement of particles in liquid suspension according to the Brownian motion. The trajectory of the particles within a certain volume is recorded by a camera that captures the scattered light as the laser illuminates the particles. NTA software collects the data on multiple particles simultaneously. The concentration and diameter of the particles can be calculated by the Stokes-Einstein equation. Therefore, NTA allows direct observation of EVs or exosomes in solution. So far, two modes of NTA are developed for exosome identification and analysis, including conventional and fluorescent NTA (fl-NTA).
Advantages of NTA-based exosome identification
- Simpler sample processing
- Direct analysis in solution
- Faster detection speed
- Better guarantees the original state of exosomes
A better understanding of the quality of your exosome samples
When you choose NTA-based exosome identification of Creative Proteomics, you can get a better understanding of the quality of your exosome samples in a time- and labor-saving manner. We provide conventional and fl-NTA to reliably determine the physical properties of exosome preparations and to determine their concentrations. Conventional NTA allows accurate measurement of exosome concentration and diameter. While fl-NTA allows further in-depth analysis of exosome size, count, and phenotype. There are two protocols used for exosome analysis by fl-NTA, including the use of the general cell membrane dyes and the antibody-based methods. We have been providing exosome research services and solutions for years. Our experienced and skillful experts can help determine exosome enrichment and allow the comparison of sample phenotypes. Specifically, we accept a variety of sample types, such as plasma, urine, saliva, and so on.
Features of our services
- Get better NTA data on exosomes
- The skillful and experienced scientific team
- End-to-end service
- Best after-sale service
Reliable and accurate identification of exosomes will contribute to eventual translatability into disease diagnosis, drug delivery, therapy, and vaccines. Creative Proteomics is a leading custom service provider in the field of subcellular and single-cell research. We are committed to providing innovative solutions to power your biological research. If you have a question or want general information, please feel free to contact us. We are happy to help you.
Reference
- Bachurski, Daniel, et al. "Extracellular vesicle measurements with nanoparticle tracking analysis-An accuracy and repeatability comparison between NanoSight NS300 and ZetaView." Journal of extracellular vesicles 8.1 (2019): 1596016.
* For Research Use Only. Not for use in diagnostic procedures.