Chloroplast Proteomics Services
Online InquiryChloroplasts are places where plants reduce and assimilate carbon dioxide and form carbohydrates through photosynthesis. It is also the place where plant amino acids, fatty acids and terpenoids are synthesized and where nitrite and sulfate are reduced. The number of chloroplast proteins accounts for 10% to 25% of its total protein. Most of the proteins involved in the special biochemical synthesis and metabolic processes in the chloroplast are highly segmented and localized, forming a specific subproteome of each region.
Creative Proteomics provides a one-stop service for chloroplast proteomics, which aims to conduct a comprehensive analysis of chloroplast from a holistic perspective. The main contents include: 1) Analysis of plant chloroplast biogenesis and structure function; 2) Analysis of plant chloroplast's photosynthetic response regulation mechanism in different environments; 3) Analysis of subproteome information of different membrane regions and membrane compartment microregions; 4) Analysis of the role of chloroplast subproteins in metabolic networks.
We Provide the Following Services, including but not Limited to:
Study chloroplast proteins
1. Chlorophyll inner and outer envelope proteins
Due to the semi-autonomous nature of chloroplasts, most of its proteins are encoded by nuclear genes and transported into chloroplasts after cytoplasmic synthesis. Therefore, the chloroplast envelope transport system plays a vital role.
- Study the transport complexes involved in transporting nuclear-encoded proteins
- Study the chloroplast localization input pathway
- Study the membrane proteins involved in the transport of ions and cell metabolites
- Study the regulatory mechanism in the synthesis and assembly of chloroplast-specific membrane lipid components
- Confirm the distribution and function of key proteases on the capsule
- Study the relationship between chloroplast envelope and antioxidant system
- Analyzing the relationship between the chloroplast envelope and its genomic DNA replication and transcription
- Study the interaction of chloroplast membrane proteins with DNA or RNA
2. Chloroplast matrix protein
The chloroplast matrix is the key site for photosynthetic carbon assimilation and anabolic metabolism of many substances in the chloroplast, and most of the protein components involved in the transcription and translation of the chloroplast genome are also distributed in it.
- Isolation and purification of matrix proteins
- Study of proteolytic systems in the form of protein complexes in the chloroplast matrix
- Study of the location and transfer of chloroplast matrix proteins
Study thylakoid protein
Thylakoids are a key component of chloroplasts. Photosynthesis and most photosynthesis-related proteins and protein complexes are located in thylakoid membranes and thylakoid cavities.
1. Thylakoid membrane protein
- Study on the composition, structure and function of PSII complex
- Study on the composition, structure and function of PSI complex
- Study on structure and function of Cytb6f complex
- Study on the structure and function of ATP synthase
2. Thylakoid lumen protein
- Study on the localization and function of thylakoid cavity proteins
Advantages of the Chloroplast Proteomics Service:
- High performance mass spectrometry system: equipped with Q Exactive Hybrid Quadrupole-Orbitrap and Agilent 6540 Q-TOF high performance mass spectrometer with multiple mass spectrometry techniques such as SRM, PRM and SWATH full scan collection, which effectively improving the identification and quantification of low abundance subcellular related proteins.
- Multi-database integration: by integrating multiple related high-quality protein databases, the accuracy and flux of subcellular protein localization can be greatly improved.
- Multi-dimensional data analysis: in addition to the comparison with the more complete related subcellular database, multiple software was used for the prediction analysis of subcellular localization based on the calculation method to further increase the identification flux.
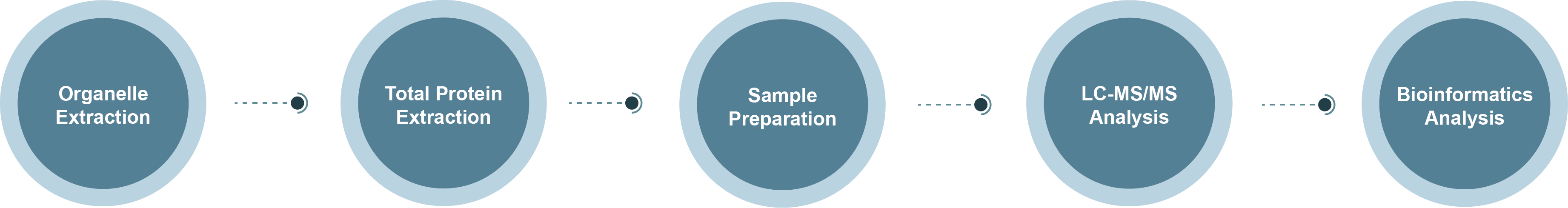
Chloroplast Proteomics Technology Roadmap
Data Analysis of Chloroplast Proteomics
| 1. Data output statistics and quality control | 2. Protein identification analysis (Density map, Scatter plot, Venn diagram) | 3. Quantitative protein analysis (Density map, Scatter plot, Venn diagram) |
| 4. Protein GO analysis | 5. Protein pathway analysis | 6. Protein COG analysis |
| 7. GO enrichment analysis of differential proteins | 8. Pathway enrichment analysis of differential proteins | 9. COG analysis of differential proteins |
References
- Moyet L, Salvi D, et al. Preparation of Membrane Fractions (Envelope, Thylakoids, Grana, and Stroma Lamellae) from Arabidopsis Chloroplasts for Quantitative Proteomic Investigations and Other Studies//Plant Membrane Proteomics. Humana Press, New York, NY, 2018: 117-136.
- Dogra V, Kim C. Chloroplast protein homeostasis is coupled with retrograde signaling. Plant signaling & behavior, 2019, 14(11): 1656037.
- Zhang S, Zhang H, et al. The caseinolytic protease complex component CLPC1 in Arabidopsis maintains proteome and RNA homeostasis in chloroplasts. BMC plant biology, 2018, 18(1): 192.
Related Services
* For Research Use Only. Not for use in diagnostic procedures.