Golgi Apparatus Proteomics Services
Online InquiryGolgi apparatus plays a critical role in post-translational modification and secretory pathway, and participates in various cellular activities, including apoptosis, mitosis, stress response, etc. The alterations, mis-localization, and changes in modification and secretion of its proteins have been found in process of a range of diseases, including cancer, Parkinson's disease, and so forth.
Creative Proteomics provides a one-stop service for Golgi apparatus proteomics, whose purpose is to perform a comprehensive analysis of Golgi apparatus. The main contents include: 1) analysis of dynamic changes in Golgi protein composition, expression level, secretion, and posttranslational modification; 2) subcellular localization of proteins; 3) identifying both soluble and membrane proteins; 4) determination of the protein function in Golgi apparatus; 5) analysis of interaction partners. Golgi apparatus proteomics provides fundamental information for uncovering novel membrane proteins and posttranslational modification, protein secretory pathway, improved insights into Golgi apparatus function, interactions and cellular mechanisms, further exploring the pathogenesis of diseases associated with Golgi apparatus, and promoting development of targeted therapies.
We Provide the Following Services, including but not Limited to:
Separation and purification of nucleoli
- Sucrose density barrier
- Discontinuous sucrose gradient
Study on the function and dynamic changes of Golgi apparatus proteins
- Study the location of Golgi apparatus proteins
- Study the function of Golgi apparatus protiens
- Investigate the protein secretory pathway
Qualitative and quantitative analysis of Golgi apparatus protein
Proteins are isolated using a rapid protocol based on differential centrifugation, and these proteins are subjected to mass spectrometry to identify Golgi apparatus proteins. Creative Proteomics provides differential analysis of the identified proteins, and quantitative analysis of proteins. According to your specific project, different quantification strategies will be applied.
Analysis of posttranslational modification of proteins
Golgi apparatus proteins are analyzed by Multidimensional protein identification technology (MudPIT), which combines a cation exchange pre-fractionation and RP HPLC separation of tryptic peptides. Creative Proteomics provides posttranslational modification analysis of proteins by using modified MudPIT.
Bioinformatics analysis
For example, Golgi apparatus protein function analysis, homology analysis and evolution analysis.
Advantages of the Golgi Proteomics Services:
- Efficient and stable protein extraction method: according to your sample types, we have optimized Golgi apparatus isolation protocol and protein extraction scheme, and increased the protein flux and extraction purity.
- MudPIT establishes more comprehensive organellar proteomes and has been optimized for analyzing covalent modifications and membrane proteins.
- High performance mass spectrometry system: equipped with Q Exactive Hybrid Quadrupole-Orbitrap and Agilent 6540 Q-TOF high performance mass spectrometer with multiple mass spectrometry techniques such as SRM, PRM and SWATH full scan collection, which effectively improving the identification and quantification of low abundance subcellular related proteins.
- Multi-database integration: the accuracy and flux of subcellular protein determination and localization can be largely improved by integrating multiple related high-quality protein databases
- Multi-dimensional data analysis: to further increase the identification flux, complete related subcellular database and multiple software will be used for the prediction analysis of subcellular localization.
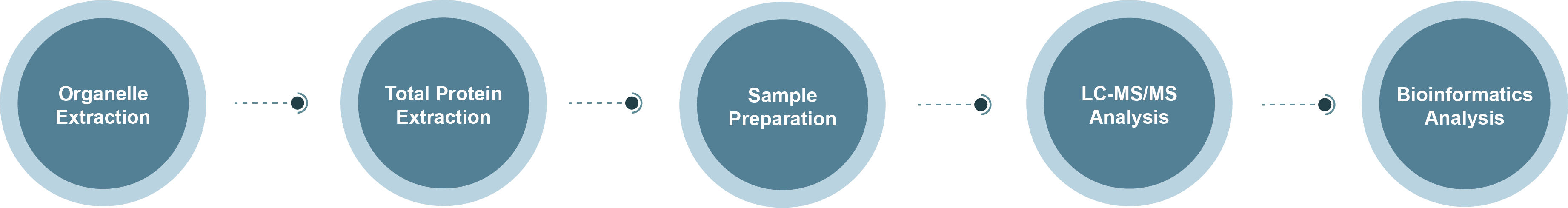
Golgi Proteomics Technology Roadmap
Data Analysis of Golgi Proteomics
| 1. Data output statistics and quality control | 2. Protein identification analysis (Density map, Scatter plot, Venn diagram) | 3. Quantitative protein analysis (Density map, Scatter plot, Venn diagram) | 4. Protein GO analysis |
| 5. Protein pathway analysis | 6. Protein COG analysis | 7. GO enrichment analysis of differential proteins | 8. Pathway enrichment analysis of differential proteins |
| 9. COG analysis of differential proteins | 10. Posttranslational modification analysis |
References
- Smirle J., Au C.E., Jain M., et al. Cell Biology of the Endoplasmic Reticulum and the Golgi Apparatus through Proteomics. Cold Spring Harb Perspect Biol. 2013;5:a015073
- Bexiga M.G. and Simpson J.C. Human Diseases Associated with Form and Function of the Golgi Complex. Int J Mol Sci. 2013;14(9):18670–18681.
- Hansen S.F., Eber B., Rautengarten C., Heazlewood J.L.(2016) Proteomic Characterization of Golgi Membranes Enriched from Arabidopsis Suspension Cell Cultures. In: Brown W. (eds) The Golgi Complex. Methods in Molecular Biology, vol 1496. Humana Press, New York, NY
- Morre D.J., Keenan T.W., and Morre D.M.Golgi apparatus isolation and use in cell-free systems. Protoplasma (1993) 172:12-26
- Wu C.C., MacCoss M.J., Mardones,G., et al. Organellar Proteomics Reveals Golgi Arginine Dimethylation. Molecular Biology of the Cell. 2004 Jun; 15(6): 2907–2919.
Related Services
* For Research Use Only. Not for use in diagnostic procedures.