Complexome Profiling Service
Online InquiryMany proteins are accustomed to other biomolecules (e.g., proteins, nucleic acids, carbohydrates, or lipids) to stabilize or dynamically bind to form macromolecular assemblies to carry out their functions. Protein complexes can be transient or stable, depending on the type of biological process involved, and often require additional factors to orchestrate the assembly of individual subunits into fully-fledged functional macromolecular entities. Conservative interaction sites allow competitive docking of different proteins, which allows proteins to bind to different interaction partners at the same binding site to form unique complexes with different functions. Changes in the cellular environment will likely cause dynamic changes in protein interactions, leading to the formation and remodeling of protein complexes.
When the interaction of the components of a protein complex is impaired and its normal formation is affected, the associated cellular processes may be compromised and lead to biological dysfunction. Altered stability and dynamics of protein complexes are often associated with the development and progression of disease. Identification of components involved in molecular actions through interaction proteomics can help to study protein function or dysfunction in disease states and to gain more insight into pathological mechanisms.
Creative Proteomics offers complexome profiling services. Collects information on the entire interactome in biological samples through an untargeted proteomic strategy that does not require specific antibodies or tags for enrichment. Mass spectrometry is used to obtain protein interaction data for natural protein complexes and to provide additional information about their natural mass, stoichiometry and recent protein turnover in protein complexes. The method is suitable for studying protein complexes, studying the dynamic processes of complex assemblies, remodeling and protein turnover.
Currently, complexome profiling is widely used to explore the composition and abundance of mitochondrial oxidative phosphorylation (OXPHOS) complexes in several species and to identify novel protein interactors involved in their assembly, maintenance and function.
The Workflow of Complexome Profiling Service
Complex analysis combines graded separation of protein complexes under natural conditions by electrophoresis, size exclusion chromatography or density gradient centrifugation. This is followed by peptide digestion and quantitative tandem MS identification. Resulting data are analyzed by clustering to visualize the inventory, abundance and alignment of multi-protein complexes in biological samples.
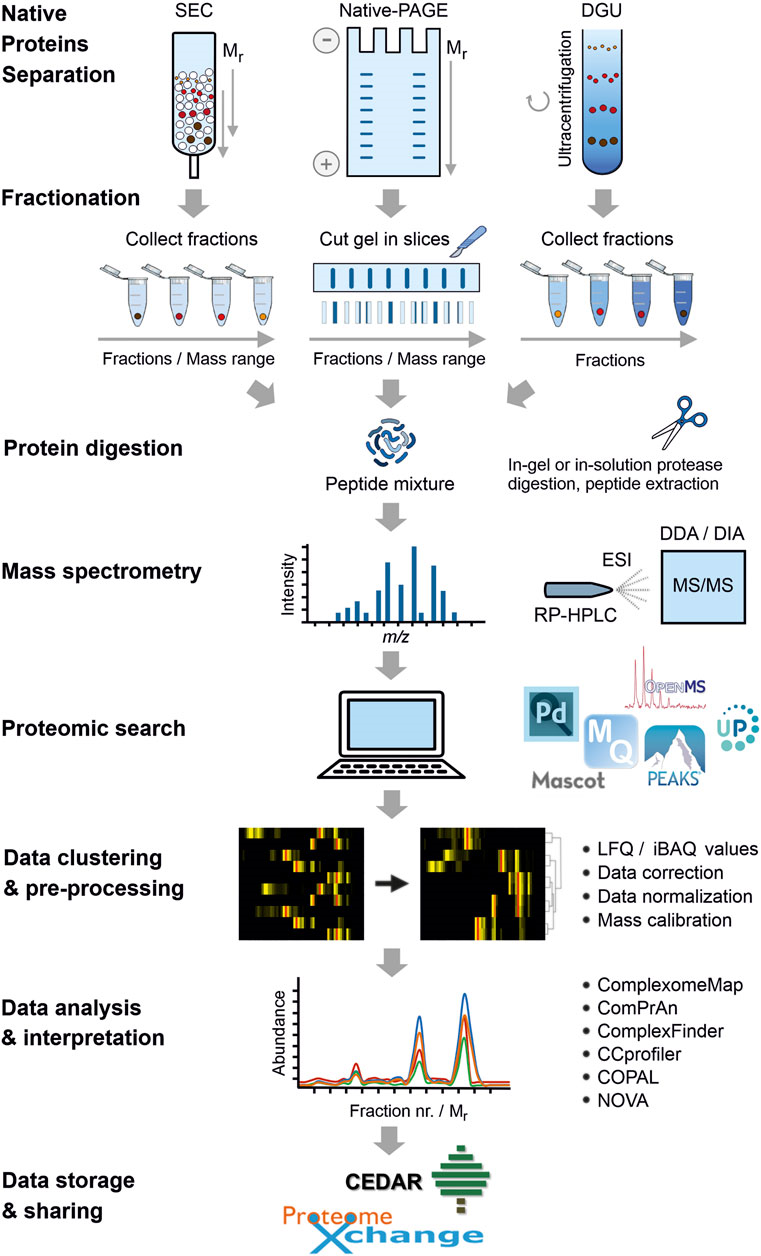 Overall workflow of complexome profiling (Cabrera et al., 2021)
Overall workflow of complexome profiling (Cabrera et al., 2021)
References
- Cabrera-Orefice, A., Potter, A., et al. (2021). Complexome Profiling-Exploring Mitochondrial Protein Complexes in Health and Disease. Frontiers in Cell and Developmental Biology, 9, 796128-796128.
- Wittig, I., & Malacarne, P. F. (2021). Complexome profiling: Assembly and remodeling of protein complexes. International Journal of Molecular Sciences, 22(15), 7809.
* For Research Use Only. Not for use in diagnostic procedures.