Chloroplast Analysis Services
Online InquiryChloroplast is a semi-autonomous organelle that performs photosynthesis commonly found in land plants, algae and some protozoa, and consists of 3 parts: chloroplast membrane, cystoid and stroma. It is the site of numerous different chemical reactions and its main function is to perform photosynthesis, in addition to being involved in the biosynthesis of amino acids and fatty acids. Chloroplasts have been the subject of extensive research efforts trying to understand the molecular mechanisms that regulate their biochemical capacity, development and evolutionary origin. The rise of subcellular histology has provided a powerful approach to elucidate the constituent proteomic and metabolomic studies of chloroplasts and has contributed more to the understanding of chloroplast function.
Creative Proteomics provides you with a professional chloroplast multi-omics analysis platform. We have high-resolution mass spectrometry instruments and a professional bioinformatics team to provide you with chloroplast proteomic, chloroplast metabolomic and chloroplast lipidomic analysis services. We aim to provide you with high-quality services to accelerate the progress of your project and open up new horizons.
The Chloroplast Assays We Offer Include but Are Not Limited to:
1. Chloroplast isolation and purification
The chloroplasts are centrifuged by percoll density gradient in order to isolate intact unbroken chloroplasts. The chloroplasts are then lysed using cell lysis solution to isolate chloroplast proteins.
2. Chloroplast proteomics analysis
We are able to realize chloroplast proteomes of different plant species under different stress conditions, such as A. thaliana, pea, barley, zucchini, sugar beet, rice, and wheat.
- Study of protein expression patterns: study of the characterization of all proteins in a particular cell or tissue under specific conditions
- Study of protein function patterns: reveal all protein functions and their modes of action
- Study of protein-protein and protein-DNA interactions
- Structural analysis of proteins
- Analysis of chloroplast outer and inner membrane proteins
- Analysis of post-translational modifications of chloroplast proteins
3. Chloroplast metabolomics analysis
- Chloroplast metabolite differential expression analysis
- Qualitative and quantitative analysis of chloroplast metabolites
4. Chloroplast lipidomic analysis
- Analysis of chloroplast lipid composition and levels
- Analysis of differential expression of chloroplast lipid molecules
- Qualitative and quantitative analysis of chloroplast lipid molecules
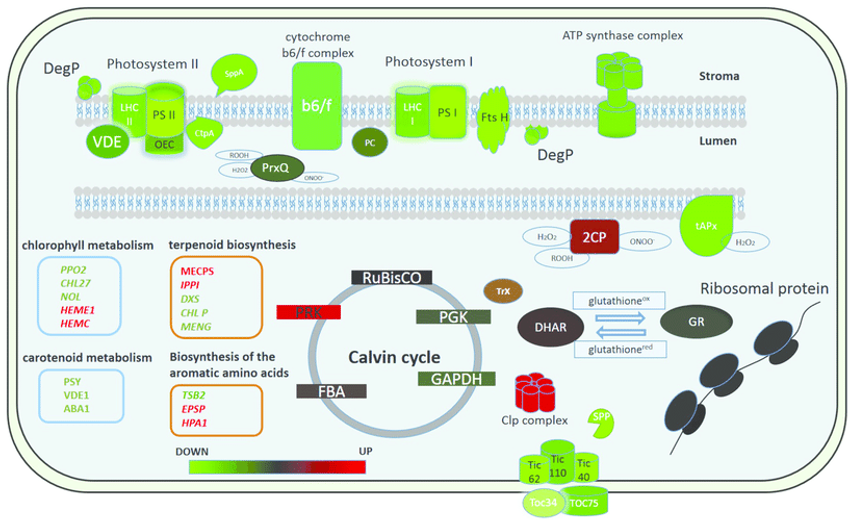 Putative model of the chloroplast proteome changes in response to short-term complex stress in P. patens (Fesenko et al., 2016).
Putative model of the chloroplast proteome changes in response to short-term complex stress in P. patens (Fesenko et al., 2016).
Basic Analysis
- Data quality control and molecular identification
- Expression quantitative analysis
- Differential analysis
- Functional annotation and enrichment analysis
Applications of Chloroplast Analysis
- Protein studies of genetic diversity: detecting changes in genetic diversity by comparing the proteome with the metabolome
- Proteomic study of mutants: The study of protein expression changes caused by gene mutations by histological methods can reveal the mechanisms of some plant physiological and ecological processes.
- Abiotic environmental factor proteomics study: In the plant survival environment, some abiotic factor stresses, such as drought, salinity, chilling, ozone, hypoxia and mechanical damage, can have serious effects on plant growth, development and survival. These stresses can cause a large number of protein/metabolite changes in kind and expression. The proteome or metabolome allows us to better understand the mechanisms of injury by abiotic stresses and the mechanisms of plant adaptation to the abiotic environment.
- Proteomic studies of biotic environmental factors: plant growth and development are not only closely related to abiotic factors, but also greatly influenced by biotic factors such as animals, plants, and microorganisms.
References
- de Luna-Valdez, L. A., León-Mejía, P., et al. (2015). Chloroplast omics. In PlantOmics: The Omics of Plant Science (pp. 533-558). Springer, New Delhi.
- Fesenko, I., Seredina, A., Arapidi, G., et al. (2016). The Physcomitrella patens chloroplast proteome changes in response to protoplastation. Frontiers in plant science, 7, 1661.
* For Research Use Only. Not for use in diagnostic procedures.