Cell Surface Proteomics Service
Online Inquiry- Service Details
- Case Study
The collection of exposed plasma membrane proteins, collectively referred to as the surface group, is involved in several important cellular processes, such as communication between cells and their surroundings and regulation of transport across the lipid bilayer. The surfaceome also plays a key role in the immune system through recognition and presentation of antigens, with possible malfunctions associated with disease. Surface proteins have been explored as potential cellular markers, disease biomarkers and therapeutic drug targets.
Compared with intracellular proteins, the low efficiency of extracting surface proteins from hydrophobic membranes and aqueous media and the low abundance of surface proteins hinder the analysis of surface proteins. Creative Proteomics has endeavored to investigate and address these issues by developing a platform for cell surface protein analysis.
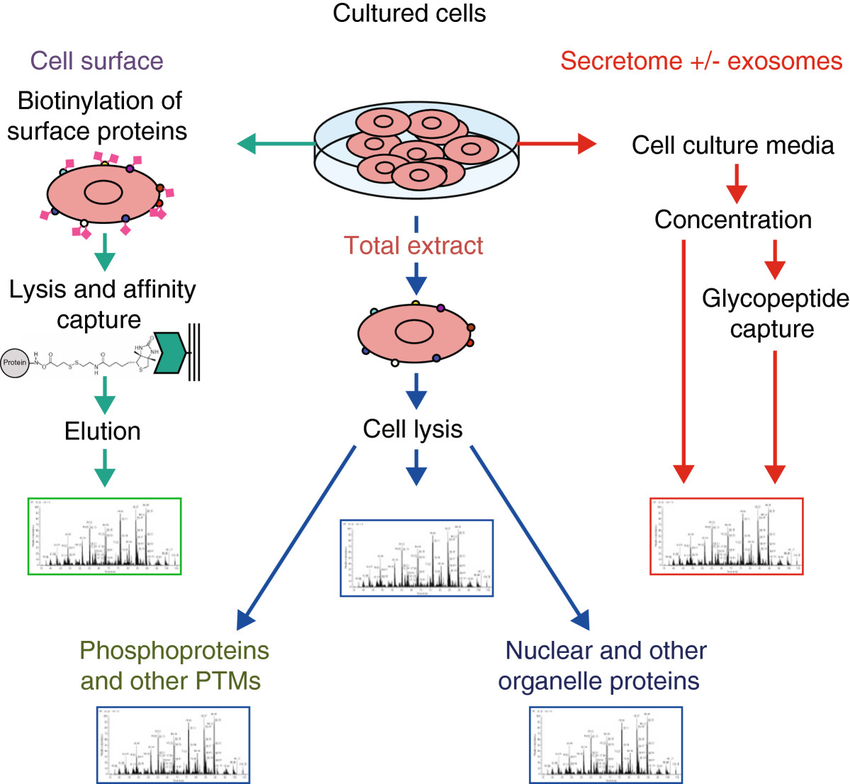 Analysis of the proteome of cell populations by sub-compartment (Hanash et al., 2014).
Analysis of the proteome of cell populations by sub-compartment (Hanash et al., 2014).
What is Included in Our Cell Surface Proteomics Analysis?
Identification and Quantification of Cell Surface Proteins: Utilization of advanced Mass Spectrometry (MS) techniques to identify and quantify cell surface proteins, resulting in a detailed catalog of these proteins and their respective abundance.
Protein Expression and Localization: Assessment of the expression levels of cell surface proteins, along with information about their localization within the cell membrane, shedding light on their spatial distribution.
Protein-Protein Interaction Analysis: Investigation of protein-protein interactions at the cell surface through immunoprecipitation (IP) and Co-immunoprecipitation (Co-IP), providing critical insights into complex signaling networks governing cellular functions.
Post-Translational Modification Characterization: In-depth exploration of post-translational modifications (PTMs) on cell surface proteins, including glycosylation, phosphorylation, and more, significantly influencing protein functions.
Functional Insights: Conducting functional assays to decipher the roles of cell surface proteins in various cellular processes, essential for the design of targeted therapeutic strategies.
Cell Surface Proteomics Analytical Techniques
Mass Spectrometry (MS): We utilize state-of-the-art mass spectrometers, including high-resolution instruments such as the Thermo Fisher Orbitrap series and the AB Sciex TripleTOF series, in combination with nano-liquid chromatography (nano-LC) systems. The use of nano-LC enhances the sensitivity and resolution of the analysis. This approach allows for the precise identification and quantification of cell surface proteins, even in complex samples. Nano-LC-MS is particularly effective for analyzing low-abundance proteins and post-translational modifications.
Western Blotting: Our Western Blotting services utilize gel electrophoresis systems (e.g., Bio-Rad Mini-PROTEAN) and transfer equipment (e.g., Bio-Rad Trans-Blot Turbo) to separate and transfer proteins. The proteins are then detected using chemiluminescence or fluorescence-based imaging systems (e.g., Bio-Rad ChemiDoc).
Immunoprecipitation (IP) and Co-immunoprecipitation (Co-IP): We perform IP and Co-IP experiments using magnetic bead-based separation systems such as Dynabeads. These techniques are commonly carried out with microcentrifuges and magnetic racks, as well as specialized antibodies to target specific cell surface proteins.
Biotinylation Assays: For biotinylation, we employ biotin labeling kits and streptavidin-based affinity purification systems. The assays are often performed in microcentrifuge tubes with centrifugation equipment.
Sample Requirements for Cell Surface Proteomics
| Sample Requirement | Details |
|---|---|
| Sample Type | Cell membrane or cell surface proteins extracted from target cells. |
| Sample Quantity | Minimum of 1 x 10^6 cells. |
| Sample Purity | Highly purified cell surface proteins, free from cytoplasmic or intracellular contaminants. |
| Sample Preparation | Gentle extraction methods to preserve the integrity of cell surface proteins. |
| Sample Storage | -80°C for long-term storage, avoid freeze-thaw cycles. |
| Labeling and Tagging | If applicable, provide information on any labeling or tagging of proteins. |
| Protein Concentration | Minimum 2 μg/μl, with a total of at least 50 μg of protein. |
| Buffer Composition | Describe the buffer used for protein extraction and storage. |
| Sample Integrity Assessment | Agarose gel electrophoresis to check for degradation or contamination. |
| Sample Documentation | Provide detailed information on the cell type, source, and any relevant experimental conditions. |
| Delivery Conditions | -80°C on dry ice for shipping, with proper documentation and sample tracking. |
Applications of Cell Surface Proteomics
Precision Drug Target Discovery: Identify and quantify cell surface proteins for precise drug target discovery, enhancing therapeutic outcomes.
Biomarker Identification: Unearth disease biomarkers for early detection and monitoring of conditions like cancer, cardiovascular diseases, and infectious ailments.
Immunology and Vaccines: Investigate immune responses and support the development of effective vaccines by understanding the interaction between cell surface proteins, immune cells, and pathogens.
Cancer Research: Advance cancer research by exploring cancer-specific markers, elucidating tumor diversity, and aiding in the development of targeted therapies.
Neuroscience: Gain insights into neuronal communication, synaptic plasticity, and neurodegenerative diseases through the study of neurotransmitter receptors and synaptic proteins.
Stem Cell Research: Characterize and sort stem cells based on surface markers for applications in regenerative medicine and tissue engineering.
Microbial Insights: Study the surface proteins of bacteria, viruses, and pathogens to develop innovative antimicrobial strategies.
Molecular Pathways: Uncover complex protein-protein interactions and signaling pathways at the cell surface to decipher cellular processes and regulatory mechanisms.
Functional Profiling: Understand the functions of cell surface proteins to design targeted therapeutic strategies and interventions.
With the help of powerful bioinformatics technology, our technicians have rich analytical experience in the field of bioinformatics and are skilled in using various bioinformatics analysis tools to deeply analyze and mine the data, which can realize the screening of potential biomarkers.
Reference
- Hanash, S., & Schliekelman, M. (2014). Proteomic profiling of the tumor microenvironment: recent insights and the search for biomarkers. Genome medicine, 6(2), 1-12.
Case: Identification of Cell Surface Proteins Associated with Colorectal Adenoma-to-Carcinoma Progression
Background
Colorectal cancer (CRC) is a significant health concern, and there is a need for specific biomarkers that can distinguish low-risk adenomas from high-risk adenomas and CRCs. Cell surface proteins are of particular interest for potential targeting in molecular imaging and cancer treatment. This study aims to identify cell surface proteins associated with the progression of colorectal adenomas to carcinomas.
Samples
The study utilized CRC cell lines, both chromosomally unstable and microsatellite instability (MSI) cell lines, for the identification of cell surface proteins. These cell lines served as models to explore potential biomarkers.
Technical Methods
Protein Isolation: Extracellular domains of membrane proteins were biotinylated in the selected CRC cell lines, followed by cell lysis. This process allowed for the separation of biotinylated cell surface protein fractions from non-biotinylated intracellular protein fractions.
Western Blotting: To validate the cell surface protein isolation procedure, Western blotting was performed for E-cadherin, a cell adhesion molecule. The results confirmed the enrichment of E-cadherin within cell surface fractions compared to intracellular fractions in the cell lines.
Proteomic Analysis: The isolated cell surface protein fractions were subjected to 1D gel electrophoresis and stained with Coomassie Brilliant Blue. These fractions were further processed for protein identification using nanoLC-MS/MS. A total of 2609 proteins were identified in the cell surface fractions across the five CRC cell lines.
Candidate Biomarker Selection: To identify potential cell surface biomarkers associated with adenoma-to-carcinoma progression, several selection criteria were applied. Proteins detected in at least four out of five cell lines, with predicted transmembrane domains and elevated mRNA levels in carcinomas compared to adenomas, were considered as candidate biomarkers. This approach yielded a list of 44 potential markers.
Validation: Two of the candidate biomarkers, GLUT1 and PrPC, were selected for further validation. Their presence on the cell surface of CRC cell lines was confirmed through fluorescence-activated cell sorting analysis. Immunohistochemistry was performed on tissue microarrays (TMAs) containing colon adenomas and carcinomas to validate their increased expression in CRC.
Results
The proteomic analysis identified 2609 proteins in the cell surface fractions of CRC cell lines, with 44 potential cell surface biomarkers selected for adenoma-to-carcinoma progression.
Validation of two candidate biomarkers, GLUT1 and PrPC, confirmed their increased expression in CRC compared to adenomas.
Classification of adenomas into low-risk and high-risk categories based on DNA copy number alterations revealed that high-risk adenomas and carcinomas displayed higher expression levels of GLUT1 and PrPC.
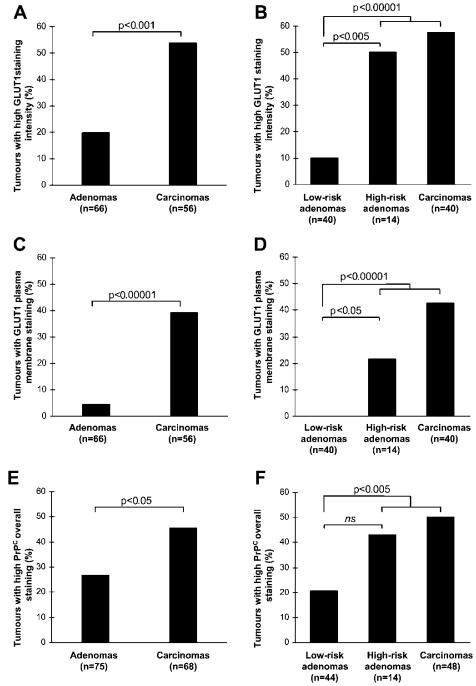 Quantification of glucose transporter type 1 (GLUT1) and prion protein (PrPC) immunohistochemical staining of TMAs
Quantification of glucose transporter type 1 (GLUT1) and prion protein (PrPC) immunohistochemical staining of TMAs
Reference
- De Wit, Meike, et al. "Cell surface proteomics identifies glucose transporter type 1 and prion protein as candidate biomarkers for colorectal adenoma-to-carcinoma progression." Gut 61.6 (2012): 855-864.
* For Research Use Only. Not for use in diagnostic procedures.