Subcellular Structure & Organelle Proteomics Solution
Online Inquiry- Service Details
- Case Study
What is Subcellular Structure & Organelle Proteomics?
Subcellular structure & organelle proteomics is a specialized branch of proteomics dedicated to exploring the hidden realms within cells. It peels back the layers of cellular complexity to reveal the diverse subcellular compartments and organelles that orchestrate life at the molecular level.
At Creative Proteomics, we believe that to truly understand the cell, one must venture into its deepest recesses. Subcellular structure & organelle proteomics is the key that unlocks this complexity. It's about:
Characterization and Localization
We decipher the identities and precise locations of proteins within specific subcellular niches. From the nucleus to mitochondria, endoplasmic reticulum to the Golgi apparatus, our expertise allows us to pinpoint proteins and their functions with accuracy.
Dynamics and Functional Insights
It's not just about who's where; it's about understanding the dynamic changes within these structures. Creative Proteomics excels in capturing shifts in protein composition, expression levels, and post-translational modifications. This information offers invaluable insights into the functional dynamics of subcellular compartments.
Subcellular structure & organelle proteomics goes beyond scientific exploration; it's the gateway to understanding the very essence of cellular function. It dissects the molecular machinery responsible for cellular processes, paving the way for breakthroughs in fields ranging from medicine to biotechnology.
Our Specialized Proteomics Services
1. Mitochondrial Proteomics Services
We can analyze the dynamic changes of mitochondrial protein composition, expression level and modification status. Clarify the expression and function patterns of all proteins in mitochondria. Analyze the interaction of mitochondrial proteins with other molecules.
2. Chloroplast Proteomics Services
We can analyze the biogenesis and structural function analysis of plant chloroplasts, the sub-proteome information of different membrane regions and membrane regions, and the role of chloroplast subproteins in the metabolic network.
3. Nuclear Proteomics Services
We can carry out the separation and purification of nuclear protein and nuclear protein, and study the expression, structure and function of nuclear protein. We also provide more precise nucleolar proteomics services and nuclear pore complex proteomics services.
4. Peroxisome Proteomics Services
We have had experience in studying the biogenesis of peroxisomes, the function of peroxisome membrane proteins and matrix proteins, and the relationship between peroxisomes and other diseases.
5. Plasma Membrane Proteomics Services
We can provide cell plasma membrane enrichment, cell plasma membrane protein extraction, separation and identification, and plasma membrane proteome related research.
6. Golgi Apparatus Proteomics Services
We can provide Golgi protein localization research, functional research and protein secretion pathway research, etc.
7. Endoplasmic Reticulum Proteomics Services
We can provide research on the interaction between endoplasmic reticulum and plasma membrane, mitochondria, Golgi apparatus, endocytosis, lipid droplets and peroxisomes, endoplasmic reticulum protein purification and quantitative qualitative analysis
8. Centrosome Proteomics Services
We can provide quantitative and qualitative research on centrosome protein, centrosome replication regulation, etc. Explore the relationship between the centrosome and various physiological functions and the relationship between the centrosome and disease.
9. Exosome Proteomics Services
We can provide exosomes separation and purification, identification, exosomal protein function model analysis, protein post-translational modification analysis, etc.
Subcellular Structure & Organelle Analytical Techniques
Our proteomics services rely on a range of high-precision mass spectrometry instruments, including:
- Thermo Scientific Orbitrap Fusion Lumos Tribrid Mass Spectrometer: This exceptional instrument combines quadrupole, Orbitrap, and linear ion trap technologies, providing high-resolution and high-accuracy mass spectrometry. It enables precise identification and quantification of proteins within subcellular compartments.
- Waters SYNAPT G2-Si High Definition Mass Spectrometry System: With its innovative ion mobility technology, this mass spectrometer offers advanced separation capabilities. It enhances our ability to capture dynamic changes in protein composition and post-translational modifications within subcellular structures.
- Bruker MALDI-TOF/TOF Mass Spectrometer: This instrument excels in matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) analysis. It is particularly valuable for mapping the precise locations of proteins within subcellular compartments, shedding light on their functional roles.
What Our Proteomics Service Can Provide?
1. Organelle Isolation and Protein Purification
The separation of organelles and the separation and purification of subcellular structural proteins are important steps for studying specific intracellular structures, organelles, proteins or evaluating the interconnection between these macromolecular structures. We can provide you with the classification and separation of common organelles and the separation and purification of subcellular structural proteins, such as mitochondria, chloroplasts, nucleoli, etc.
2. Subcellular Structures Protein Identification
Subcellular protein identification analysis is the use of mass spectrometry to identify all or specific proteins in cells or protein gels/liquids after separation, purification, and enrichment, so as to fully understand the types of proteins in specific environments.
3. Quantitative Analysis of Subcellular Proteome
We can provide you with subcellular protein quantitative analysis. Detect rich changes in protein. Combine differential analysis with quantitative techniques to help study the changes or functions of the proteome under certain physiological or pathological conditions.
4. Post-Translational Modification Analysis
We can provide post-translational modification characterization and protein modification sites determination services. Discovering and studying protein modification sites can promote our understanding of protein function.
5. Protein Subcellular Localization
Knowing the subcellular location of proteins is helpful to the study of gene function, protein interaction and mechanism of action. We can provide subcellular location services, including immunofluorescence labeling, fluorescent protein fusion and subcellular location prediction.
6. Subcellular Protein-Protein Interaction Analysis
Each protein does not independently perform its assigned function in the cell. It usually interacts with other proteins to form large complexes that perform specific functions in a specific time and space. Moreover, the functions of some proteins can only be performed after the complex is formed. We can provide protein interactions analysis service to accelerate your project.
7. Subcellular Protein Structure Analysis
We can provide you with subcellular protein structure analysis services, including X-ray single crystal diffraction service, cryo-electron microscopy service and nuclear magnetic resonance spectrum service.
Applications of Subcellular Structure & Organelle Proteomics
Cell Biology: Elucidating the dynamics of cellular processes and organelle interactions.
Neuroscience: Investigating neuronal subcellular compartments for neurodegenerative diseases.
Cancer Biology: Identifying subcellular protein alterations in cancer cells.
Drug Discovery: Validating drug targets within specific organelles.
Immunology: Understanding immune cell organelles in infection and autoimmunity.
Bioinformatics Data Analysis for Subcellular Structure & Organelle Proteomics
| Bioinformatics Data Analysis Components | Description |
|---|---|
| Quantitative Proteomics Analysis | - Protein Abundance: Quantify protein levels within subcellular compartments, enabling relative expression comparisons. - Differential Expression: Identify significantly altered proteins using statistical methods, aiding in biomarker or target discovery. - Hierarchical Clustering: Organize proteins into clusters with similar expression patterns to unveil functional groups. |
| Post-translational Modification (PTM) Analysis | - PTM Identification: Identify and characterize various PTMs (e.g., phosphorylation, glycosylation) to unravel regulatory mechanisms. - Site Localization: Pinpoint the exact amino acid residues modified by PTMs for precise functional insights. |
| Functional Analysis | - Gene Ontology (GO) Enrichment: Categorize proteins based on biological processes, molecular functions, and cellular components, revealing functional roles. - Functional Annotation: Annotate proteins with known functions to explore their involvement in specific cellular processes or diseases. - Pathway Enrichment: Identify enriched biological pathways among subcellular proteins to understand functional context. |
| Interaction Analysis | - Protein-Protein Interaction (PPI) Analysis: Build protein-protein interaction networks to uncover key players and pathways within subcellular structures. |
| Subcellular Localization Prediction | - Subcellular Localization Prediction: Predict the specific subcellular compartments where proteins are likely to reside, enhancing functional understanding. |
| Differential Gene Expression Analysis | - Identification of Differentially Expressed Genes: Analyze gene expression data to identify genes showing significant expression changes within subcellular compartments. |
| Biomarker Discovery | - Identification of Potential Biomarkers: Explore the dataset for proteins that may serve as potential biomarkers for diseases or biological processes. |
| Multi-Omics Integration | - Integration with Other Omics Data: Combine subcellular proteomics data with other omics data types, such as genomics or metabolomics, for a comprehensive systems biology approach. |
| Subcellular Protein Distribution Visualization | - Data Visualization: Utilize visualization tools to create subcellular protein distribution maps, enhancing the understanding of protein spatial localization. |
The subcellular structure or content you want to analyze is not within the above range? Please contact us. Our professionals will proactively contact you after receiving your email to understand your needs and provide you with assistance.
Case. Proteomic Profiling of Host Cellular Responses to HCMV Infection: Unveiling Spatial-Temporal Dynamics and Organelle Alterations
Background
Human Cytomegalovirus (HCMV) is a pathogenic virus that infects human cells, leading to various cellular responses. To better understand how HCMV affects cellular organelles and their functions, a multidisciplinary study was conducted using live cell microscopy, mass spectrometry-based proteomics, and functional analyses.
Samples
The study utilized MRC5 human lung fibroblasts as the cellular model. These cells were infected with the HCMV strain AD169, and samples were collected at various time points (24, 48, 72, 96, and 120 hours post-infection) to cover the entire viral infectious cycle. Uninfected cells were also sampled at these time points for comparison.
Technical Methods
Sample Collection and Preparation: Cellular samples were collected at various time points following HCMV infection (24, 48, 72, 96, and 120 hours) alongside uninfected control samples. These samples were meticulously prepared to extract proteins for analysis.
Protein Extraction: A standardized protocol was followed for the extraction of total protein content from both infected and uninfected cell samples. This process ensured the capture of a broad range of cellular proteins.
Quantitative Proteomics: Advanced mass spectrometry-based techniques, particularly liquid chromatography-tandem mass spectrometry (LC-MS/MS), were employed for quantitative proteomic analysis. This methodology allowed for the simultaneous quantification of numerous proteins with high precision.
Differential Protein Expression Analysis: Robust computational analysis was conducted to identify differentially expressed proteins (DEPs) by comparing the protein abundance levels between infected and uninfected samples. Statistical analyses were applied to ascertain the significance of these changes.
Functional Annotation: DEPs were functionally annotated to classify them based on their biological functions, subcellular localizations, and involvement in cellular pathways. This annotation provided crucial insights into the functional roles of these proteins during the host cell's response to HCMV infection.
Temporal Dynamics: The proteomic data were meticulously examined over time, enabling the observation of dynamic alterations in protein abundance during the progression of HCMV infection. This temporal analysis revealed the evolving host cellular responses.
Subcellular Localization Predictions: Machine learning models were harnessed to predict the subcellular localization of proteins based on their quantitative profiles. This prediction helped uncover the translocation of proteins to different cellular compartments upon infection.
Validation: Selected DEPs were experimentally validated using techniques such as Western blotting and immunofluorescence assays. These validation experiments confirmed the changes in protein abundance observed through proteomics analysis.
Identification of Viral Proteins: In addition to host proteins, the proteomics approach also identified viral proteins expressed during HCMV infection, contributing to the understanding of host-virus interactions at the molecular level.
Construction of Protein-Protein Interaction Networks: Interaction networks were constructed to explore how DEPs interacted with one another and to elucidate potential roles these proteins played in the cellular responses to HCMV infection.
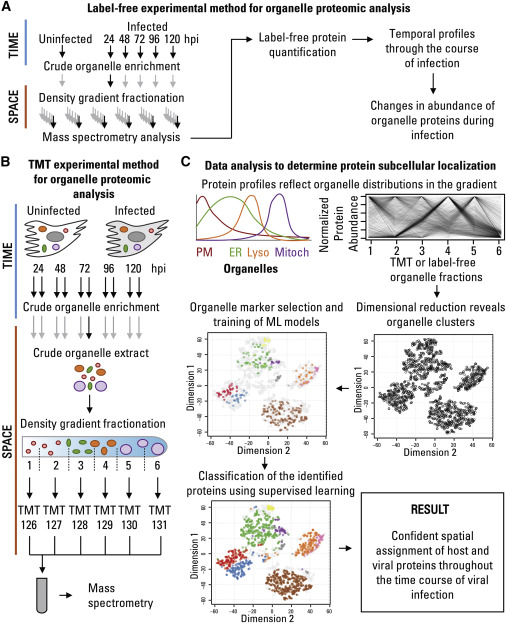 Proteomic Approach to Define Spatial-Temporal Changes in Organelle Proteins throughout HCMV Infection
Proteomic Approach to Define Spatial-Temporal Changes in Organelle Proteins throughout HCMV Infection
Results
Identification of Differentially Expressed Proteins (DEPs): The proteomics analysis identified a significant number of differentially expressed proteins (DEPs) during HCMV infection compared to uninfected controls. These DEPs were categorized based on their functions and subcellular localizations.
Temporal Changes in Protein Abundance: The study revealed dynamic changes in the abundance of specific proteins over time post-infection (at 24, 48, 72, 96, and 120 hours). This temporal analysis provided insights into the progression of cellular responses to HCMV infection.
Alterations in Organelle Composition: Through organelle fractionation and proteomics, the researchers observed alterations in the composition of cellular organelles. This included changes in the abundance of organelle-specific marker proteins, suggesting that HCMV infection had a profound impact on organelle function and organization.
Functional Enrichment Analysis: Functional enrichment analysis of DEPs was performed to gain insights into the biological processes and pathways affected by HCMV infection. This analysis highlighted specific cellular functions that were significantly modulated during infection, such as immune response pathways and metabolic processes.
Protein Localization Predictions: Machine learning models were employed to predict the subcellular localization of proteins based on their quantitative profiles. This allowed for the identification of proteins that translocated to different organelles upon HCMV infection. Such translocations provided insights into the virus's manipulation of host cell machinery.
Validation of Proteomics Findings: To validate the proteomics results, selected DEPs were further analyzed using techniques like Western blotting and immunofluorescence. These validation experiments confirmed the changes in protein abundance observed in the proteomics analysis.
Identification of Viral Proteins: In addition to host cell proteins, the study also identified viral proteins expressed during HCMV infection. This information was critical for understanding how the virus interacts with host cell machinery.
Interaction Networks: Protein-protein interaction networks were constructed to elucidate the interactions between DEPs and their potential roles in cellular responses to HCMV infection. This network analysis provided a systems-level view of the host-virus interaction.
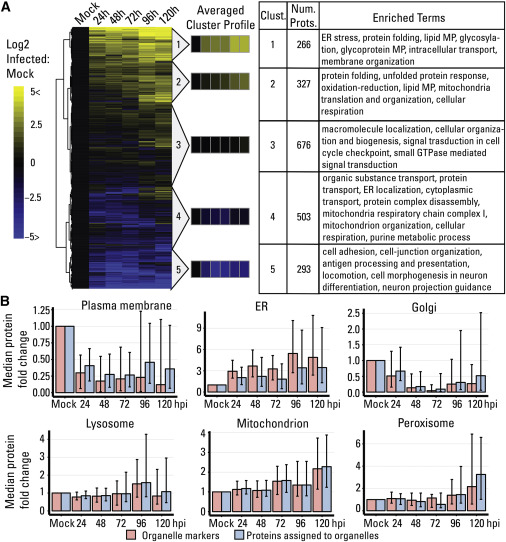 Temporal Label-free Proteomic Analysis of Organelles throughout HCMV Infection
Temporal Label-free Proteomic Analysis of Organelles throughout HCMV Infection
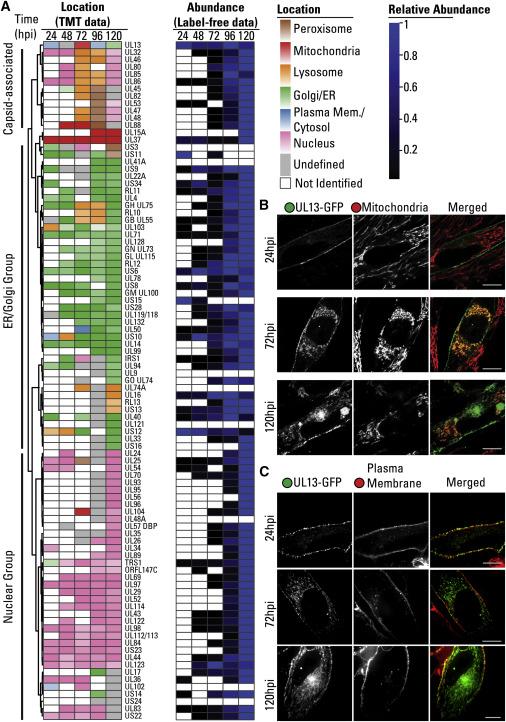 Temporal Localizations and Abundances of Viral Proteins throughout Infection
Temporal Localizations and Abundances of Viral Proteins throughout Infection
Reference
- Beltran, Pierre M. Jean, Rommel A. Mathias, and Ileana M. Cristea. "A portrait of the human organelle proteome in space and time during cytomegalovirus infection." Cell systems 3.4 (2016): 361-373.
* For Research Use Only. Not for use in diagnostic procedures.